(Written for release of v6.0, June 2020)
| Expand | ||
|---|---|---|
| ||
|
...
| Note | ||
|---|---|---|
| ||
It is recommended that you familiarise yourself with R first by sitting our Introduction to R tutorial. It also requires that you have the DataSHIELD training environment installed on your machine, see our Installation Instructions for Linux, Windows, or Mac. |
| Tip | ||
|---|---|---|
| ||
DataSHIELD support is freely available in the DataSHIELD forum by the DataSHIELD community. Please use this as the first port of call for any problems you may be having, it is monitored closely for new threads. DataSHIELD bespoke user support and also user training classes are offered on a fee-paying basis. Please enquire at datashield@newcastle.ac.uk for current prices. |
Introduction
This tutorial introduces users to DataSHIELD commands and syntax. Throughout this document we refer to R, but all commands are run in the same way in Rstudio. This tutorial contains a limited number of examples; further examples are available in each DataSHIELD function manual page that can be accessed via the help function.
...
| Tip | ||
|---|---|---|
| ||
DataSHIELD commands call functions that range from carrying out pre-requisite tasks such as login to the datasources, to generating basic descriptive statistics, plots and tabulations. More advance functions allow for users to fit generalized linear models and generalized estimating equations models. R can list all functions available in DataSHIELD. This section explains the functions we will call during this tutorial. Although this knowledge is not required to run DataSHIELD analyses it helps to understand the output of the commands. It can explain why some commands call functions that return nothing to the user, but rather store the output on the server of the data provider for use in a second function. In DataSHIELD the person running an analysis (the client) uses client-side functions to issue commands (instructions). These commands initiate the execution (running) of server-side functions that run the analysis server-side (behind the firewall of the data provider). There are two types of server-side function: assign functions and aggregate functions. Assign functions do not return an output to the client, with the exception of error or status messages. Assign functions create new objects and store them server-side either because the objects are potentially disclosive, or because they consist of the individual-level data which, in DataSHIELD, is never seen by the analyst. These new objects can include:
Assign functions return no output to the client except to indicate an error or useful messages about the object store on server-side. Aggregate functions analyse the data server-side and return an output in the form of aggregate data (summary statistics that are not disclosive) to the client. The help page for each function tells us what is returned and when not to expect an output on client-side. |
...
Please follow instructions to Start the Opal VMs.
Recall from the installation instructions, the Opal web interface:
...
| Anchor | ||||
|---|---|---|---|---|
|
Start R/RStudio
Load Packages
- The following relevant R packages are required for analysis:
...
| Code Block | ||||
|---|---|---|---|---|
| ||||
#load libraries library(DSI) library(DSOpal) library(dsBaseClient) |
Build your login dataframe
| Tip | ||
|---|---|---|
| ||
The DataSHIELD cloud training environment does not use fixed IP addresses, the client and opal training server addresses change each training session. As part of the user tutorial you learn how to build a DataSHIELD login dataframe. In a real world instance of DataSHIELD this is populated with secure certificates not text based usernames and passwords. |
Login Dataframe
- Build login dataframe.
| Code Block | ||||
|---|---|---|---|---|
| ||||
builder <- DSI::newDSLoginBuilder()
builder$append(server = "study1", url = "http://192.168.56.100:8080/",
user = "administrator", password = "datashield_test&",
table = "CNSIM.CNSIM1", driver = "OpalDriver")
builder$append(server = "study2", url = "http://192.168.56.101:8080/",
user = "administrator", password = "datashield_test&",
table = "CNSIM.CNSIM2", driver = "OpalDriver")
logindata <- builder$build() |
...
| Anchor | ||||
|---|---|---|---|---|
|
Assign individual variables on login
Users can specify individual variables to assign to the server-side R session. It is best practice to first create a list of the Opal variables you want to analyse.
...
| Div | ||
|---|---|---|
| ||
Basic statistics and data manipulations
Descriptive statistics: variable dimensions and class
It is possible to get some descriptive or exploratory statistics about the assigned variables held in the server-side R session such as number of participants at each data provider, number of participants across all data providers and number of variables. Identifying parameters of the data will facilitate your analysis.
...
| Code Block | ||
|---|---|---|
| ||
Aggregated (exists("D")) [=============================================================] 100% / 0s
Aggregated (classDS("D$LAB_HDL")) [====================================================] 100% / 1s
$study1
[1] "numeric"
$study2
[1] "numeric" |
Descriptive statistics: quantiles and mean
As LAB_HDL is a numeric variable the distribution of the data can be explored.
...
| Code Block | ||
|---|---|---|
| ||
Aggregated (meanDS(D$LAB_HDL)) [=======================================================] 100% / 0s
$Mean.by.Study
EstimatedMean Nmissing Nvalid Ntotal
study1 1.569416 360 1803 2163
study2 1.556648 555 2533 3088
$Nstudies
[1] 2
$ValidityMessage
ValidityMessage
study1 "VALID ANALYSIS"
study2 "VALID ANALYSIS" |
Descriptive statistics: assigning variables
So far all the functions in this section have returned something to the screen. Some functions (assign functions) create new objects in the server-side R session that are required for analysis but do not return an anything to the client screen. For example, in analysis the log values of a variable may be required.
...
| Code Block | ||
|---|---|---|
| ||
Aggregated (meanDS(LAB_HDL.c)) [=======================================================] 100% / 0s
$Mean.by.Study
EstimatedMean Nmissing Nvalid Ntotal
study1 0.007416316 360 1803 2163
study2 -0.005352231 555 2533 3088
$Nstudies
[1] 2
$ValidityMessage
ValidityMessage
study1 "VALID ANALYSIS"
study2 "VALID ANALYSIS"
|
Generating contingency tables
The function ds.table creates contingency tables of a categorical variables. The default is set to run on pooled data from all studies, to obtain an output of each study set the argument type='split' .
...
| Code Block | ||||
|---|---|---|---|---|
| ||||
$chisq.tests $chisq.tests$chisq.test_TABLE.STUDY.1_counts Pearson's Chi-squared test with Yates' continuity correction data: input.array.source.specific X-squared = 3.8767, df = 1, p-value = 0.04896 $chisq.tests$chisq.test_TABLE.STUDY.2_counts Pearson's Chi-squared test with Yates' continuity correction data: input.array.source.specific X-squared = 3.5158, df = 1, p-value = 0.06079 $chisq.tests$chisq.test_TABLES.COMBINED_all.sources_counts Pearson's Chi-squared test with Yates' continuity correction data: combine.array.all.sources X-squared = 7.9078, df = 1, p-value = 0.004922 $validity.message [1] "Data in all studies were valid" |
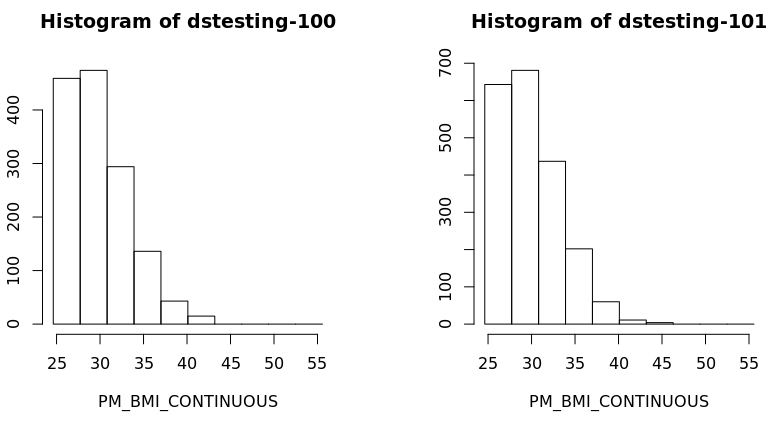
Generating graphs
It is currently possible to produce 4 types of graphs in DataSHIELD: histograms, contour plots, heatmap plots and scatter plots.
| Anchor | ||||
|---|---|---|---|---|
|
Histograms
| Tip | ||
|---|---|---|
| ||
In the default method of generating a DataSHIELD histogram outliers are not shown as these are potentially disclosive. The text summary of the function printed to the client screen informs the user of the presence of classes (bins) with a count smaller than the minimal cell count set by data providers. Note that a small random number is added to the minimum and maximum values of the range of the input variable. Therefore each user should expect slightly different printed results from those shown below: |
...
| Anchor | ||||
|---|---|---|---|---|
|
Contour plots
Contour plots are used to visualize a correlation pattern.
...
| Anchor | ||||
|---|---|---|---|---|
|
Heat map plots
An alternative way to visualise correlation between variables is via a heat map plot.
...
| Tip |
|---|
The functions |
Saving Graphs / Plots in R Studio
- Any plots will appear in the bottom right window in R Studio, within the
plottab - Select
export>save as image
...
- The plot will now be accessible from your Home folder directory structure.
Sub-setting
| Tip | ||
|---|---|---|
| ||
Sub-setting is particularly useful in statistical analyses to break down variables or tables of variables into groups for analysis. Repeated sub-setting, however, can lead to thinning of the data to individual-level records that are disclosive (e.g. the statistical mean of a single value point is the value itself). Therefore, DataSHIELD does not subset an object below the minimal subset length set by the data providers (typically this is ≤ 4 observations). |
...
- ds.subsetByClass
- ds.dataFrameSubset
- ds.subset (WARNING: this function will be deprecated in the release of 6.1, all functionallity has been added to ds.dataFrameSubset which will become the one-stop replacement).
| Anchor | ||||
|---|---|---|---|---|
|
Sub-setting using ds.subsetByClass
- The
ds.subsetByClassfunction generates subsets for each level of acategoricalvariable. If the input is a data frame it produces a subset of that data frame for each class of each categorical variable held in the data frame. - Best practice is to state the categorical variable(s) to subset using the
variablesargument, and the name of the subset data using thesubsetsargument. - The example subsets
GENDERfrom our assigned data frameD, the subset data is namedGenderTables:
...
| Anchor | ||||
|---|---|---|---|---|
|
Sub-setting using ds.subset
| Warning |
|---|
This function is soon to be deprecated. Its replacement will be ds.dataFrameSubset(). ds.dataFrameSubset() uses very different arguments to ds.subset() |
...
| Code Block | ||
|---|---|---|
| ||
ds.histogram('BMI25plus$PM_BMI_CONTINUOUS', datasources = opals)
Warning: dstesting-100: 2 invalid cells
Warning: dstesting-101: 1 invalid cells
[[1]]
$breaks
[1] 23.93659 27.17016 30.40373 33.63731 36.87088 40.10445 43.33803 46.57160 49.80518 53.03875 56.27232
$counts
[1] 365 511 331 150 49 15 0 0 0 0
$density
[1] 0.079212771 0.110897880 0.071834047 0.032553194 0.010634043 0.003255319 0.000000000 0.000000000 0.000000000 0.000000000
$mids
[1] 25.55337 28.78695 32.02052 35.25409 38.48767 41.72124 44.95482 48.18839 51.42196 54.65554
$xname
[1] "xvect"
$equidist
[1] TRUE
attr(,"class")
[1] "histogram"
[[2]]
$breaks
[1] 23.93659 27.17016 30.40373 33.63731 36.87088 40.10445 43.33803 46.57160 49.80518 53.03875 56.27232
$counts
[1] 506 750 476 229 62 11 4 0 0 0
$density
[1] 0.0767450721 0.1137525773 0.0721949690 0.0347324536 0.0094035464 0.0016683711 0.0006066804 0.0000000000 0.0000000000 0.0000000000
$mids
[1] 25.55337 28.78695 32.02052 35.25409 38.48767 41.72124 44.95482 48.18839 51.42196 54.65554
$xname
[1] "xvect"
$equidist
[1] TRUE
attr(,"class")
[1] "histogram |
Modelling
Horizontal DataSHIELD allows the fitting of generalised linear models (GLM). In the GLM function the outcome can be modelled as continuous, or categorical (binomial or discrete). The error to use in the model can follow a range of distribution including gaussian, binomial, Gamma and poisson. In this section only one example will be shown, for more examples please see the manual help page for the function.
| Anchor | ||||
|---|---|---|---|---|
|
Generalised linear models
- The function
ds.glmis used to analyse the outcome variableDIS_DIAB(diabetes status) and the covariatesPM_BMI_CONTINUOUS(continuous BMI),LAB_HDL(HDL cholesterol) andGENDER(gender), with an interaction between the latter two. In R this model is represented as:
...
| Tip | ||
|---|---|---|
| ||
After every iteration in the glm, each study returns non disclosive summaries (a score vector and an information matrix) that are combined on the client R session. The model is fitted again with the updated beta coefficients, this iterative process continues until convergence or the maximum number of iterations is reached. The output of |
Conclusion
| Tip |
|---|
You have now completed our basic DataSHIELD training. Some extended practicals will be coming soon. Also remember you can:
|
...