The answers and suggested code below are for the extended practical session that allows you to implement basic DataSHIELD functions and interpret the results.
# load libraries
library(opal)
library(dsBaseClient)
server <- c("study1", "study2", "study3")
url <- c("http://XXXXXX:8080")
table <- c("DASIM.DASIM1", "DASIM.DASIM2", "DASIM.DASIM3")
logindata <- data.frame(server, url, user="administrator", password="datashield_test&", table)
# login and assign the whole dataset
opals <- datashield.login(logins=logindata, assign=TRUE) |
# Check the levels of categorigal BMI (1=normal, 2=overweight, 3=obese)
ds.levels('D$PM_BMI_CATEGORICAL')
# Check the levels of gender (0=males, 1=females)
ds.levels('D$GENDER')
# Create a subset dataset that includes only the obese people
ds.subset(x='D', subset='BMI_3', logicalOperator='PM_BMI_CATEGORICAL==', threshold=3)
# See how many obese people are in each study
ds.dim('BMI_3')
# Create a subset dataset that includes only the obese males
ds.subset(x='BMI_3', subset='BMI_3_males', logicalOperator='GENDER==', threshold=0)
# Check how many obese males are in each study
ds.dim('BMI_3_males')
# Calculate the global mean and global variance of continuous bmi for obese males
ds.mean('BMI_3_males$PM_BMI_CONTINUOUS', type = 'combine')
ds.var('BMI_3_males$PM_BMI_CONTINUOUS', type = 'combine') |
The global mean and the global variance of BMI are 33.04723 and 6.134642 respectively. |
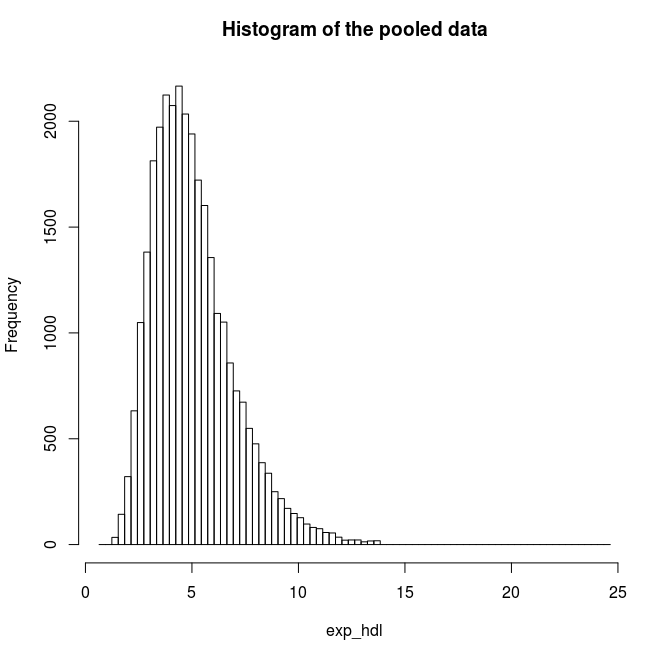
Find the quantile mean and plot a histogram of pooled data for the exponent and for the logarithm of LAB_HDL measurement.
# Assign a new variable which gives the exponents of HDL
ds.exp(x='D$LAB_HDL', newobj='exp_hdl')
# Find the quantile mean of the exponents of HDL
ds.quantileMean('exp_hdl')
# Plot a histogram for the exponents of HDL
ds.histogram('exp_hdl') |
Quantiles of the pooled data 5% 10% 25% 50% 75% 90% 95% Mean 2.555388 2.922673 3.660593 4.727446 6.072125 7.653894 8.725955 5.066809 |

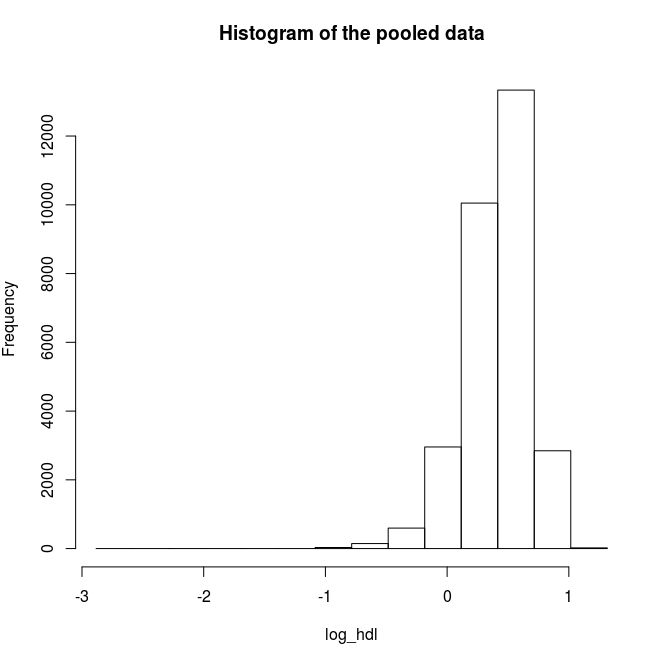
# Assign a new variable which gives the logarithms of HDL
ds.log(x='D$LAB_HDL', newobj='log_hdl')
# Find the quantile mean of the logarithms of HDL
ds.quantileMean('log_hdl')
# Plot a histogram for the logarithms of HDL
ds.histogram('log_hdl') |
Quantiles of the pooled data 5% 10% 25% 50% 75% 90% -0.06384112 0.06994799 0.26052368 0.44043450 0.58983773 0.71059831 95% Mean 0.77301979 0.40754040 |
# Produce a two dimensional table for the variables GENDER and # DIS_DIAB for combined data ds.table2D(x='D$GENDER', y='D$DIS_DIAB') |
1.57% of females (pooled data) are diabetics.
|
# Produce a two dimensional table for the variables GENDER and # DIS_CVA for split data ds.table2D(x='D$GENDER', y='D$DIS_CVA', type='split') |
The percentages of males having stroke are 0.82% in study 1, 0.80% in study 2 and 0.78% in study 3.
|
|
# Apply GLM to find the linear relationship between
# LAB_GLUC_FASTING and GENDER
ds.glm("D$LAB_GLUC_FASTING ~ 1 + D$GENDER",family="gaussian") |
The relationship between glucose and gender is given by the formula: For males, GENDER=0 and therefore their average level of glucose is 4.62223776 For females, GENDER=1 their average level of glucose is 4.62223776 - 0.08929719 = 4.532941
|
# Apply GLM to find the linear relationship of LAB_GLUC_FASTING
# with GENDER and PM_BMI_CONTINUOUS
ds.glm("D$LAB_GLUC_FASTING~1+D$GENDER+D$PM_BMI_CONTINUOUS", family="gaussian") |
The level of glucose related to gender and bmi is given by the formula: While the level of bmi is increasing by one unit, the level of glucose is increasing by 0.03543909. For a female (GENDER=1) with PM_BMI_CONTINUOUS=22, the level of glucose should be
|
# clear the Datashield R sessions and logout datashield.logout(opals) |