...
Quick reminder for logging in:
| Expand |
|---|
Recall from the installation instructions, the Opal web interface is a simple check to tell if the VMs have started. Load the following urls, waiting at least 1 minute after starting the training VMs. Start R/RStudioLoad Packages| Code Block |
|---|
| #load libraries
library(DSI)
library(DSOpal)
library(dsBaseClient)
|
Build your login dataframe | Code Block |
|---|
| language | xml |
|---|
| title | Build your login dataframe |
|---|
| builder <- DSI::newDSLoginBuilder()
builder$append(server = "study1", builder <- DSI::newDSLoginBuilder()
builder$append(server = "server1", url = "httphttps://192opal-demo.168.56.100:8080obiba.org/",
user = "administratordsuser", password = "datashield_test&P@ssw0rd", table driver = "CNSIM.CNSIM1OpalDriver", driver = "OpalDriver"options='list(ssl_verifyhost=0, ssl_verifypeer=0)')
builder$append(server = "study2server2", url = "httphttps://192opal-demo.168.56.101:8080obiba.org/",
user = "administratordsuser", password = "datashield_test&P@ssw0rd", table driver = "CNSIM.CNSIM2OpalDriver", driver = "OpalDriver"options='list(ssl_verifyhost=0, ssl_verifypeer=0)')
logindata <- builder$build()
logindata <- builder$build()
connections <- DSI::datashield.login(logins = logindata, assign = TRUE)
DSI::datashield.assign.table(conns = connections, symbol = "D"DST", table = c("CNSIM.CNSIM1","CNSIM.CNSIM2")) |
| Code Block |
|---|
| DSI::datashield.logout(connections) |
|
...
| Code Block |
|---|
|
# first find the column name you wish to refer to
ds.colnames(x="DDST")
# then check which levels you need to apply a boolean operator to:
ds.levels(x="D$GENDERDST$GENDER")
?ds.dataFrameSubset |
At this stage, we want to work out what arguments are available in the DataSHIELD function so we summon the function help; the help appears as:
...
| Code Block |
|---|
|
ds.dataFrameSubset(df.name = "DDST", V1.name = "D$GENDER", V2.name = "1", Boolean.operator = "==", newobj = "CNSIM.subset.Males", datasources= connections)
ds.dataFrameSubset(df.name = "DDST", V1.name = "D$GENDER", V2.name = "0", Boolean.operator = "==", newobj = "CNSIM.subset.Females",datasources= connections) |
...
| Code Block |
|---|
|
ds.completeCases(x1="DDST",newobj="DDST_without_NA", datasources=connections) |
...
| Code Block |
|---|
|
Assigned expr. (DDST_without_NA <- completeCasesDS("D")) [================================] 100% / 1s
Aggregated (testObjExistsDS("DDST_without_NA")) [=========================================] 100% / 0s
Aggregated (messageDS("DDST_without_NA")) [===============================================] 100% / 0s
$is.object.created
[1] "A data object <D<DST_without_NA> has been created in all specified data sources"
$validity.check
[1] "<D<DST_without_NA> appears valid in all sources" |
...
| Code Block |
|---|
|
ds.dataFrameSubset(df.name = "DDST",
V1.name = "D$PMDST$PM_BMI_CONTINUOUS",
V2.name = "25",
Boolean.operator = ">=",
newobj = "subset.BMI.25.plus",
datasources = connections) |
...
| Code Block |
|---|
|
Aggregated (dataFrameSubsetDS1("DDST", "D$PM_BMI_CONTINUOUS", "25", 6, NULL, ) [==========] 100% / 1s
Assigned expr. (subset.BMI.25.plus <- dataFrameSubsetDS2("DDST", "D$PM_BMI_CONTINUOUS", "25", 6, N...
Aggregated (testObjExistsDS("subset.BMI.25.plus")) [===================================] 100% / 0s
Aggregated (messageDS("subset.BMI.25.plus")) [=========================================] 100% / 0s
$is.object.created
[1] "A data object <subset.BMI.25.plus> has been created in all specified data sources"
$validity.check
[1] "<subset.BMI.25.plus> appears valid in all sources" |
...
| Expand |
|---|
| title | Deprecated instructions on ds.subset and ds.subsetByClass, soon to be replaced with instructions using ds.dataFrameSubset |
|---|
|
In DataSHIELD there are currently 3 functions that allow us to generate subset data: - ds.subsetByClass (WARNING: this function will be deprecated in the release of 6.1, all functionality has been added to ds.dataFrameSubset which will become the one-stop replacement)
- ds.subset (WARNING: this function will be deprecated in the release of 6.1, all functionality has been added to ds.dataFrameSubset which will become the one-stop replacement).
- ds.dataFrameSubset
Sub-setting using ds.subsetByClass- The
ds.subsetByClass function generates subsets for each level of a categorical variable. If the input is a data frame it produces a subset of that data frame for each class of each categorical variable held in the data frame. - Best practice is to state the categorical variable(s) to subset using the
variables argument, and the name of the subset data using the subsets argument. - The example subsets
GENDER from our assigned data frame D , the subset data is named GenderTables :
| Code Block |
|---|
| ds.subsetByClass(x = 'DDST', subsets = "GenderTables", variables = 'GENDER', datasources = connections) |
- The output of
ds.subsetByClass is held in a list object stored server-side, as the subset data contain individual-level records. If no name is specified in the subsets argument, the default name "subClasses" is used.
| Warning |
|---|
Running ds.subsetByClass on a data frame without specifying the categorical variable in the argument variables will create a subset of all categorical variables. If the data frame holds many categorical variables the number of subsets produces might be too large - many of which may not be of interest for the analysis. |
In the previous example, the GENDER variable in assigned data frame D had females coded as 0 and males coded as 1. When GENDER was subset using the ds.subsetByClass function, two subset tables were generated for each study dataset; one for females and one for males. - The
ds.names function obtains the names of these subset data:
| Code Block |
|---|
| ds.names('GenderTables', datasources = connections)
|
| Code Block |
|---|
| Aggregated (exists("GenderTables")) [==================================================] 100% / 0s
Aggregated (classDS("GenderTables")) [=================================================] 100% / 0s
Aggregated (namesDS(GenderTables)) [===================================================] 100% / 0s
$study1
[1] "GENDER.level_0" "GENDER.level_1"
$study2
[1] "GENDER.level_0" "GENDER.level_1" |
| Anchor |
|---|
| sub_meanbyclass |
|---|
| sub_meanbyclass |
|---|
|
Sub-setting using ds.subset| Warning |
|---|
This function is soon to be deprecated. Its replacement will be ds.dataFrameSubset(). ds.dataFrameSubset() uses very different arguments to ds.subset()
Changes will be coming soon to this page. Use function help to investigate how ds.dataFrameSubset() works similarly. |
The function ds.subset allows general sub-setting of different data types e.g. categorical, numeric, character, data frame, matrix. It is also possible to subset rows (the individual records). No output is returned to the client screen, the generated subsets are stored in the server-side R session. - The example below uses the function
ds.subset to subset the assigned data frame D by rows (individual records) that have no missing values (missing values are denoted with NA ) given by the argument completeCases=TRUE . The output subset is named "D_without_NA":
| Code Block |
|---|
| ds.subset(x='DDST', subset='DDST_without_NA', completeCases=TRUE, datasources = connections)
|
| Note |
|---|
The ds.subset function prints an invalid message to the client screen to inform if missing values exist in a subset. #In order to indicate that a generated subset dataframe or vector is invalid all values within it are set to NA!
An invalid message also denotes subsets that contain less than the minimum cell count determined by data providers. |
- The second example creates a subset of the assigned data frame
D with BMI values ≥ 25 using the argument logicalOperator. The subset object is named BMI25plus using the subset argument and is not printed to client screen but is stored in the server-side R session:
| Code Block |
|---|
| ds.subset(x='DDST', subset='BMI25plus', logicalOperator='PM_BMI_CONTINUOUS>=', threshold=25, datasources = opals)
|
| Note |
|---|
The subset of data retains the same variables names i.e. column names |
- To verify the subset above is correct (holds only observations with BMI ≥ 25) the function
ds.quantileMean with the argument type='split' will confirm the BMI results for each study are ≥ 25.
| Code Block |
|---|
| ds.quantileMean('BMI25plus$PM_BMI_CONTINUOUS', type='split', datasources = opals)
$`dstesting-100`
5% 10% 25% 50% 75% 90% 95% Mean
25.3500 25.7100 27.1500 29.2000 32.0600 34.6560 36.4980 29.9019
$`dstesting-101`
5% 10% 25% 50% 75% 90% 95% Mean
25.46900 25.91800 27.19000 29.27000 32.20500 34.76200 36.24300 29.92606
|
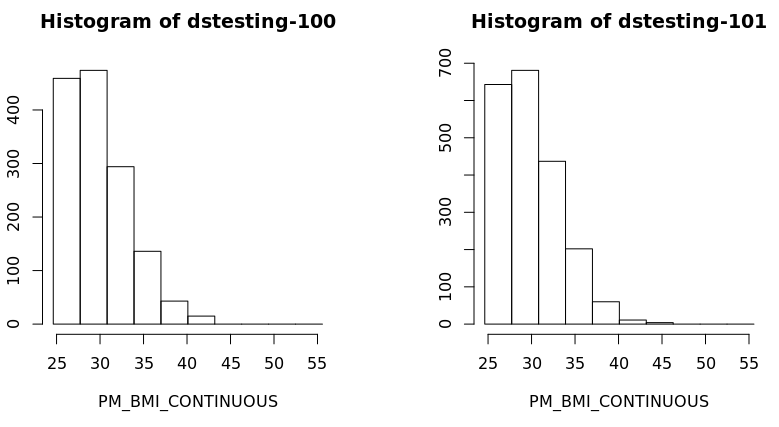
- Also a histogram of the variable BMI of the new subset data frame could be created for each study separately:
| Div |
|---|
| style | page-break-after:always; |
|---|
| ds.histogram('BMI25plus$PM_BMI_CONTINUOUS', datasources = opals) |
| Code Block |
|---|
| ds.histogram('BMI25plus$PM_BMI_CONTINUOUS', datasources = opals)
Warning: dstesting-100: 2 invalid cells
Warning: dstesting-101: 1 invalid cells
[[1]]
$breaks
[1] 23.93659 27.17016 30.40373 33.63731 36.87088 40.10445 43.33803 46.57160 49.80518 53.03875 56.27232
$counts
[1] 365 511 331 150 49 15 0 0 0 0
$density
[1] 0.079212771 0.110897880 0.071834047 0.032553194 0.010634043 0.003255319 0.000000000 0.000000000 0.000000000 0.000000000
$mids
[1] 25.55337 28.78695 32.02052 35.25409 38.48767 41.72124 44.95482 48.18839 51.42196 54.65554
$xname
[1] "xvect"
$equidist
[1] TRUE
attr(,"class")
[1] "histogram"
[[2]]
$breaks
[1] 23.93659 27.17016 30.40373 33.63731 36.87088 40.10445 43.33803 46.57160 49.80518 53.03875 56.27232
$counts
[1] 506 750 476 229 62 11 4 0 0 0
$density
[1] 0.0767450721 0.1137525773 0.0721949690 0.0347324536 0.0094035464 0.0016683711 0.0006066804 0.0000000000 0.0000000000 0.0000000000
$mids
[1] 25.55337 28.78695 32.02052 35.25409 38.48767 41.72124 44.95482 48.18839 51.42196 54.65554
$xname
[1] "xvect"
$equidist
[1] TRUE
attr(,"class")
[1] "histogram |
|
...
| Tip |
|---|
Also remember you can: |
...
