Checks for server site functions
Server side functions are like standard R scripts. It is crucial to check the code at each stages because some errors can literally cripple the package and cause it to never install on the opal server. At such stage (installation on opal) it becomes difficult to trace back the reason why the package is not 'behaving' as expected. We recommend 3 levels of checks: (1) check each line using the expected input - as you would do in standard R and double check that the output is as expected, (2) after making sure the code runs and produces the right output put the code in opal as a script and call it from the client side (3) If the code works fine as a script on opal build the package, push into GitHub, install it in opal and call it again from the client side.
Easy and fast server site function check
In the early stages of the DataSHIELD project we use to check server side functions by first building the package, pushing it into GitHub, installing the GitHub version in opal and calling the functions from the client site. This is a time consuming process particularly if you experience some issues with your remote connection to GitHub or with installing the package on opal. The second check listed below offers a much quicker way of trying out a function. Please make use of it for efficient development time management.
In this page we use the function meanDS to illustrate the 3 stage checks described above. You can practice by using the function replaceNaDS. In these checks we assume that the privacy level is set to 5 that is input objects with between 1 and 4 observations are considered invalid.
Are all the lines running without error and is the output correct?
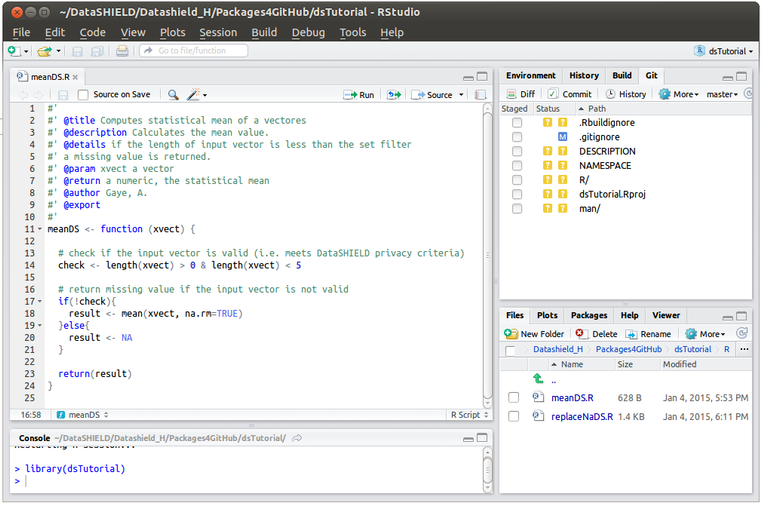
There are many ways of checking code in R, some are semi automated (e.g using the package testthat, but I am probably old school; I stick to the small scale manual and visual checks and this is what I am going to show here. As already said server side functions are standard R scripts so we should create the type(s) of inputs that the function might be provided with and see if the output corresponds to what is expected as the function's argument. The below code says it all. Go to Rstudio and on the bottom right section go to File, navigate to the R folder in your dsFirstPack and click on meanDS, this will open the script on the top left section of your Rstudio - see image below. Highlight all the code and click on Run on the top sub-menu. Our function is now loaded on the work environment and we can use it (We load the function this way because we have not built the package yet, later on it will suffice to load the package to have all its functions available in the environment.
Some checks such as the class of the vector and whether or not the vector is defined (i.e. exists) at all are done on the client side so we need not to worry about those aspects because the server site function will not be called if for example the class of the input vector is not the right one. Here we only have to make sure the code works with the expected inputs vectors.
If the function fails the checks by either (1) throwing an error at some line(s) or (2) returning an output other than the expected one; please amend the code until everything works fine.
# the function takes a 'valid' or 'invalid' numeric vector so let us generate those two vectors: # a numeric vector with 100 observations, a mean of 1 and a standard deviation of 0.5 and # a numeric vector with 4 observations, a mean of 1 and a standard deviation of 0.5 validvect <- rnorm(100, 1, 0.5) invalidvect <- rnorm (4, 1, 0.5) # pass the valid vector to the function. It should return a mean of approximately 1 meanDS(validvect) # now pass the invalid vector to the function. It should return a missing value (i.e. NA) meanDS(invalidvect)
Can the function be ran from opal as a script?
Now copy the below part of the script - make sure you do not copy the meanDS bit - the function name is specified differently in opal.
function (xvect) {
# check if the input vector is valid (i.e. meets DataSHIELD privacy criteria)
check <- isValidDS(xvect)
# return missing value if the input vector is not valid
if(!check){
result <- mean(xvect, na.rm=TRUE)
}else{
result <- NA
}
return(result)
}
- login to your opals servers (here)
- Click on the
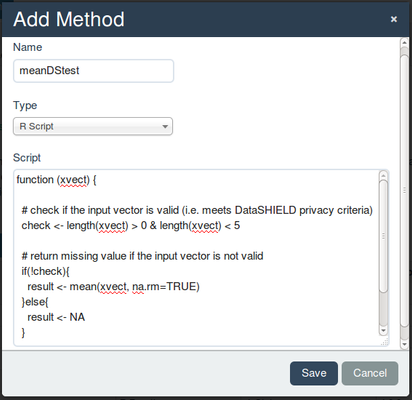
Administrationtab, then select the tabDataSHIELD. Under the sectionMethods, selectAggregatebecause we are added an aggregate function. Click on the button+Add Methodand paste the code we copied above. Add a suffix to the name to not confuse this test script with the final function if you happen to forget to delete it after testing - make sure you chooseR ScriptunderTypeas shown in the below figure and save.
- Now launch your Rstudio, load the base package for example and run for example the below code where we call the server side function as script to test it (e.g. compute the mean value of the variable 'LAB_TSC').
data(logindata) myvar <- list('LAB_TSC') opals <- datashield.login(logins=logindata,assign=TRUE,variables=myvar) datashield.aggregate(opals, meanDStest(LAB_TSC))
If everything is fine - i.e. the function computes the mean value without throwing an error - we can move on the resume the package build.
Can the function ran from opal as part of a package?
For this check we need to first build or package which is now pretty much ready. Go to this section, follow the guide to get your package to github and install it on your test servers and once you are done open your R/Rstudio load some data and call the functions to make sure they do the job just like they did when you called them from a script in the previous section.
# load the client package(s) required for the function you want to test
library(dsBaseClient)
# load the object that contains the login details
data(logindata)
# login and assign a variable of the relevant type (e.g. if your function is for a numeric vector load a numeric variable)
# By default the assigned dataset is a data frame named 'D' - so the variable will be in a data frame 'D'
myvar <- list("LAB_HDL")
opals <- datashield.login(logins=logindata,assign=TRUE,variables=myvar)
# call 'meanDS' to compute the mean of the variable 'LAB_HDL'; note how were are calling it (this is different from a using a client side function)
# we used 'datashield.aggregate' because 'meanDS' is an aggregate function.
datashield.aggregate(opals, meanDS(D$LAB_HDL))
# now let us call the function 'replaceNaDS' and replace missing values by say 0 (this is just an illustration so the replacement value may well not make sense)
# now we are calling an 'assign' function so we use 'datashield.assign' - do not forget to provide the name of the new vector which will not replace the one in 'D'
# but will rather be stored as a 'loose' vector on the server side.
datashield.assign(opals, "LAB_HDL_noNA", replaceNaDS(D$LAB_HDL, 0))
DataSHIELD Wiki by DataSHIELD is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License. Based on a work at http://www.datashield.ac.uk/wiki