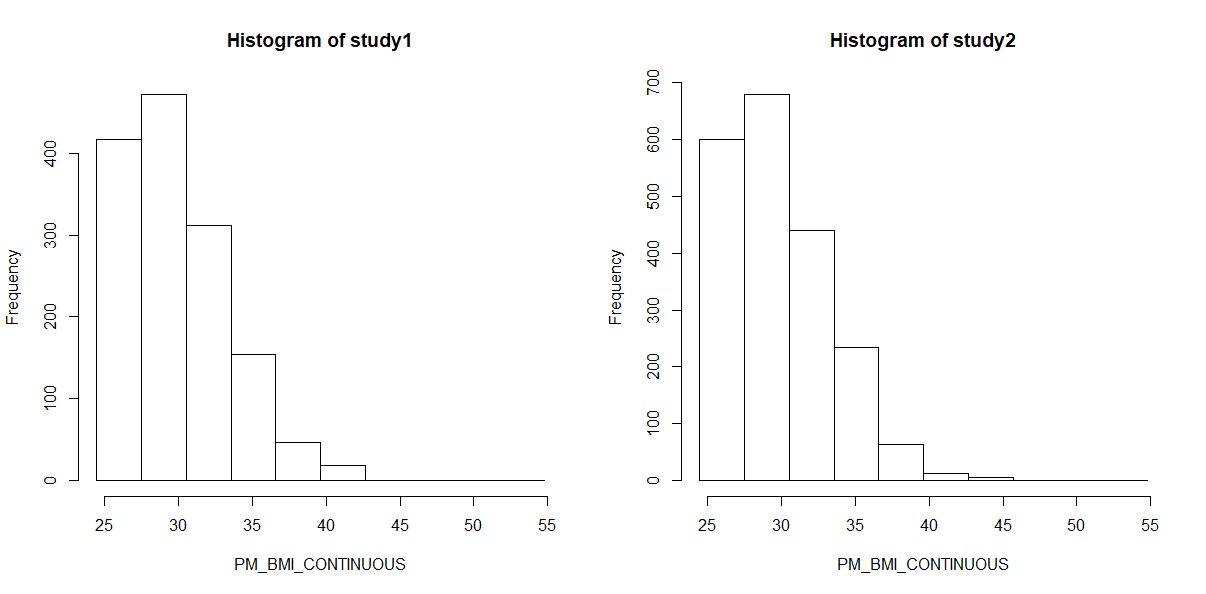DataSHIELD Training Part 5: Sub-setting
Prerequisites
It is recommended that you familiarise yourself with R first by sitting our Introduction to R tutorial.
It also requires that you have the DataSHIELD training environment installed on your machine, see our Installation Instructions for Linux, Windows, or Mac.
Help
DataSHIELD support is freely available in the DataSHIELD forum by the DataSHIELD community. Please use this as the first port of call for any problems you may be having, it is monitored closely for new threads.
DataSHIELD bespoke user support and also user training classes are offered on a fee-paying basis. Please enquire at datashield@newcastle.ac.uk for current prices.
Introduction
This is the fifth in a 6-part DataSHIELD tutorial series.
The other parts in this DataSHIELD tutorial series are:
5: Subsetting
6: Modelling
Quick reminder for logging in:
Sub-setting
Limitations on subsetting
Sub-setting is particularly useful in statistical analyses to break down variables or tables of variables into groups for analysis. Repeated sub-setting, however, can lead to thinning of the data to individual-level records that are disclosive (e.g. the statistical mean of a single value point is the value itself). Therefore, DataSHIELD does not subset an object below the minimal subset length set by the data providers (typically this is ≤ 4 observations).
In DataSHIELD there is one function that allows sub-setting of data, ds.dataFrameSubset .
You may wish to use it to:
- Subset a column of data by its "Class"
- Subset a dataframe to remove any "NA"s
- Subset a numeric column of a dataframe using a Boolean inequalilty
Sub-setting by class
You may wish to generate subsets for each level of a categorical variable. To do this we must think about which levels of that categorical variable are available, then use boolean operators to isolate them:
# first find the column name you wish to refer to ds.colnames(x="DST") # then check which levels you need to apply a boolean operator to: ds.levels(x="DST$GENDER") ?ds.dataFrameSubset
At this stage, we want to work out what arguments are available in the DataSHIELD function so we summon the function help; the help appears as:
ds.dataFrameSubset( df.name = NULL, V1.name = NULL, V2.name = NULL, Boolean.operator = NULL, keep.cols = NULL, rm.cols = NULL, keep.NAs = NULL, newobj = NULL, datasources = NULL, notify.of.progress = FALSE ) Arguments df.name a character string providing the name of the data frame to be subseted. V1.name A character string specifying the name of the vector to which the Boolean operator is to be applied to define the subset. For more information see details. V2.name A character string specifying the name of the vector to compare with V1.name. Boolean.operator A character string specifying one of six possible Boolean operators: '==', '!=', '>', '>=', '<' and '<='. keep.cols a numeric vector specifying the numbers of the columns to be kept in the final subset. rm.cols a numeric vector specifying the numbers of the columns to be removed from the final subset. keep.NAs logical, if TRUE the missing values are included in the subset. If FALSE or NULL all rows with at least one missing values are removed from the subset. newobj a character string that provides the name for the output object that is stored on the data servers. Default dataframesubset.newobj. datasources a list of DSConnection-class objects obtained after login. If the datasources the default set of connections will be used: see datashield.connections_default. notify.of.progress specifies if console output should be produced to indicate progress. Default FALSE.
So what we have learnt from this is that we must specify:
- the data frame we are working with throughout this tutorial ("D"), as the df.name argument
- the column we wish to split by class ("DST$GENDER"), as the V1.name argument
- the value we want to compare the column with, in this case a number ("0"), as the V2.name argument
- the boolean operator we want to use to compare V2.name with V1.name argument
- the specific name we want to call the new object, in string form, with the newobj argument
- as always, specify the datasources = connections
ds.dataFrameSubset(df.name = "DST", V1.name = "DST$GENDER", V2.name = "1", Boolean.operator = "==", newobj = "CNSIM.subset.Males", datasources= connections) ds.dataFrameSubset(df.name = "DST", V1.name = "DST$GENDER", V2.name = "0", Boolean.operator = "==", newobj = "CNSIM.subset.Females",datasources= connections)
Now there are two serverside objects which have split GENDER by class, to which we have assigned the names "CNSIM.subset.Males" and "CNSIM.subset.Females".
Sub-setting to remove NAs
- The example below uses the function ds.completeCases to subset the assigned data frame D
NA).The output subset is named "D_without_NA":
ds.completeCases(x1="DST",newobj="DST_without_NA", datasources=connections)
Assigned expr. (DST_without_NA <- completeCasesDS("D")) [================================] 100% / 1s
Aggregated (testObjExistsDS("DST_without_NA")) [=========================================] 100% / 0s
Aggregated (messageDS("DST_without_NA")) [===============================================] 100% / 0s
$is.object.created
[1] "A data object <DST_without_NA> has been created in all specified data sources"
$validity.check
[1] "<DST_without_NA> appears valid in all sources"
A subsequent check using ds.dim() will confirm that the new object "D_without_NA" has fewer rows than the original object "D".
Sub-set by inequality
Say we wanted to have a subset where BMI values are ≥ 25, and call it subset.BMI.25.plus
Then the V1.name argument should specify the column name for BMI, which is PM_BMI_CONTINUOUS (remember, this can always be checked by the command ds.colnames(x="D") )and the V2.name argument should specify the value to compare the column to, namely 25, using the boolean operator >=. In the DataSHIELD syntax this looks like the following:
ds.dataFrameSubset(df.name = "DST", V1.name = "DST$PM_BMI_CONTINUOUS", V2.name = "25", Boolean.operator = ">=", newobj = "subset.BMI.25.plus", datasources = connections)
The output is:
Aggregated (dataFrameSubsetDS1("DST", "DST$PM_BMI_CONTINUOUS", "25", 6, NULL, ) [==========] 100% / 1s
Assigned expr. (subset.BMI.25.plus <- dataFrameSubsetDS2("DST", "DST$PM_BMI_CONTINUOUS", "25", 6, N...
Aggregated (testObjExistsDS("subset.BMI.25.plus")) [===================================] 100% / 0s
Aggregated (messageDS("subset.BMI.25.plus")) [=========================================] 100% / 0s
$is.object.created
[1] "A data object <subset.BMI.25.plus> has been created in all specified data sources"
$validity.check
[1] "<subset.BMI.25.plus> appears valid in all sources"
The subset of data retains the same variables names i.e. column names. Note we are addressing our newly-named object on the serverside, not accessing a column of the original dataframe "DST$...." as before:
ds.colnames(x="subset.BMI.25.plus", datasources = connections)
Outputs:
Aggregated (exists("subset.BMI.25.plus")) [============================================] 100% / 0s
Aggregated (classDS("subset.BMI.25.plus")) [===========================================] 100% / 0s
Aggregated (colnamesDS("subset.BMI.25.plus")) [========================================] 100% / 0s
$study1
[1] "LAB_TSC" "LAB_TRIG" "LAB_HDL" "LAB_GLUC_ADJUSTED" "PM_BMI_CONTINUOUS"
[6] "DIS_CVA" "MEDI_LPD" "DIS_DIAB" "DIS_AMI" "GENDER"
[11] "PM_BMI_CATEGORICAL"
$study2
[1] "LAB_TSC" "LAB_TRIG" "LAB_HDL" "LAB_GLUC_ADJUSTED" "PM_BMI_CONTINUOUS"
[6] "DIS_CVA" "MEDI_LPD" "DIS_DIAB" "DIS_AMI" "GENDER"
[11] "PM_BMI_CATEGORICAL"
To verify the subset above is correct (holds only observations with BMI ≥ 25) the function
ds.quantileMean
with the argument
type='split'
will confirm the BMI results for each study are ≥ 25.
ds.quantileMean(x="subset.BMI.25.plus$PM_BMI_CONTINUOUS", type = "split", datasources= connections)
Outputs:
Aggregated (exists("subset.BMI.25.plus")) [============================================] 100% / 0s
Aggregated (classDS("subset.BMI.25.plus$PM_BMI_CONTINUOUS")) [=========================] 100% / 0s
Aggregated (quantileMeanDS(subset.BMI.25.plus$PM_BMI_CONTINUOUS)) [====================] 100% / 0s
Aggregated (lengthDS("subset.BMI.25.plus$PM_BMI_CONTINUOUS")) [========================] 100% / 0s
Aggregated (numNaDS(subset.BMI.25.plus$PM_BMI_CONTINUOUS)) [===========================] 100% / 0s
$study1
5% 10% 25% 50% 75% 90% 95% Mean
25.3500 25.7100 27.1500 29.2000 32.0600 34.6560 36.4980 29.9019
$study2
5% 10% 25% 50% 75% 90% 95% Mean
25.46900 25.91800 27.19000 29.27000 32.20500 34.76200 36.24300 29.92606
Finally we can create a histogram of these results to confirm them visually:
ds.histogram(x="subset.BMI.25.plus$PM_BMI_CONTINUOUS", datasources = connections)
Gives code output:
Aggregated (exists("subset.BMI.25.plus")) [============================================] 100% / 0s
Aggregated (classDS("subset.BMI.25.plus$PM_BMI_CONTINUOUS")) [=========================] 100% / 0s
Aggregated (histogramDS1(subset.BMI.25.plus$PM_BMI_CONTINUOUS,1,3,0.25)) [=============] 100% / 0s
Aggregated (histogramDS2(subset.BMI.25.plus$PM_BMI_CONTINUOUS,10,24.4517227951437,54.8007820545...
Warning: study1: 3 invalid cells
Warning: study2: 1 invalid cells
[[1]]
$breaks
[1] 24.45172 27.48663 30.52153 33.55644 36.59135 39.62625 42.66116 45.69606 48.73097 51.76588 54.80078
$counts
[1] 417 472 312 154 47 18 0 0 0 0
$density
[1] 0.096421960 0.109139485 0.072143049 0.035609069 0.010867703 0.004162099 0.000000000 0.000000000 0.000000000
[10] 0.000000000
$mids
[1] 25.96918 29.00408 32.03899 35.07389 38.10880 41.14371 44.17861 47.21352 50.24842 53.28333
$xname
[1] "xvect"
$equidist
[1] TRUE
attr(,"class")
[1] "histogram"
[[2]]
$breaks
[1] 24.45172 27.48663 30.52153 33.55644 36.59135 39.62625 42.66116 45.69606 48.73097 51.76588 54.80078
$counts
[1] 600 680 441 235 64 12 6 0 0 0
$density
[1] 0.0969591481 0.1098870345 0.0712649738 0.0379756663 0.0103423091 0.0019391830 0.0009695915 0.0000000000 0.0000000000
[10] 0.0000000000
$mids
[1] 25.96918 29.00408 32.03899 35.07389 38.10880 41.14371 44.17861 47.21352 50.24842 53.28333
$xname
[1] "xvect"
$equidist
[1] TRUE
attr(,"class")
[1] "histogram"
And graph:
Conclusion
The other parts in this DataSHIELD tutorial series are:
5: Subsetting
6: Modelling
Also remember you can:
- get a function list for any DataSHIELD package and
- view the manual help page individual functions
- in the DataSHIELD test environment it is possible to print analyses to file (.csv, .txt, .pdf, .png)
- take a look at our FAQ page for solutions to common problems such as Changing variable class to use in a specific DataSHIELD function.
- Get support from our DataSHIELD forum.
Related content
DataSHIELD Wiki by DataSHIELD is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License. Based on a work at http://www.datashield.ac.uk/wiki